EC for Big Learning
Join us in fusing our EC classifiers
This project is maintained by flexgp
Big Learning Activity
Have you ever wanted to run your EC algorithm in the cloud? Discouraged by the complexity of EC2? We will deploy your EC algorithm on the cloud for you with our FCUBE framework!
FCUBE supports a Bring Your Own Learner (BYOL) model: it deploys your EC algorithm to hundreds of machines and does all the data management for you. No scripts, no launch hassles, no tedious result collection. FCUBE is (EC) deployment as a service:
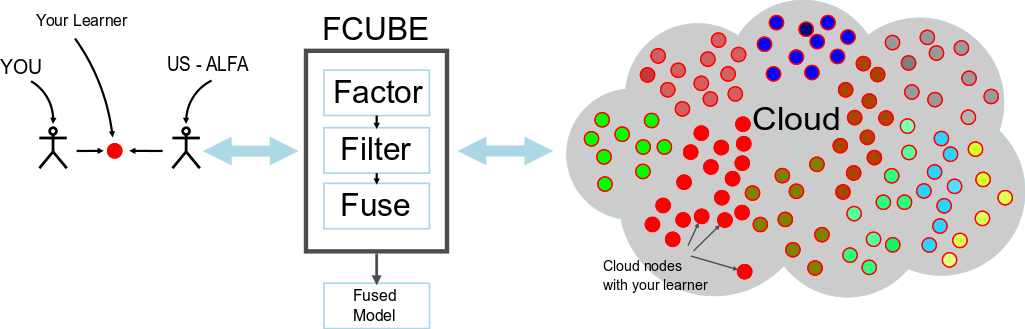
For this activity, our goal is to unite the developers of interesting EC classifier algorithms to solve relevant problems of public domain. We seek an experienced informed discussion on the various approaches and techniques without being distracted by one problem at hand. Therefore, we have set up the following format:
- Everyone gets the same computational budget in Amazon EC2
- Everyone works on the same datasets
- Organizers select the features
- You contribute a classifier learning algorithm with your own fitness function, operators and search logic
- You contribute a classifier learning algorithm which accepts training data in csv format and references a Java properties file which you provide (details below), and outputs a classifier.
- You contribute a classifier learning algorithm in executable format (Java, python) or as source code (must be compilable in Linux: C, C++ etc)
- You contribute a piece of code which applies your classifier to test data and produces labels.
- FCUBE executes your algorithm with the training data selected by the organizers
- FCUBE retrieves the solutions from the cloud nodes, computes the testing predictions, and returns them to you.
- FCUBE also filters and fuses the predictions using different methods. Everyone receives their fused results and everyone contributes to a collaborative fused solution among all contributors.
Instructions for participants
Phase 1: Bring Your Own Learner until May 31st
In a first step, you will adapt your learner to be compliant with FCUBE's interface; we provide an example learner GPFunction (one of our Java learners for numerical features) together with a split of the higgs dataset that you can use to debug yours:
Learner Training interface

Inputs:
- a path to a CSV file: we support csv files with and without headers. When headers are included, the first line of the file contains the name and type of the features (integer, float, or nominal). Check this dummy dataset with headers and without headers and this split of the Higgs dataset with headers and without headers.
- learning time deadline
- Properties File with Java syntax for your extra parameters. Check this example.
Outputs:
- a model stored in a single file on disk
Example with our GPFunction learner:
$ java -jar gpfunction.jar -train higgs_noheader_02.csv -minutes 10 -properties params.properties
Learner Predict interface
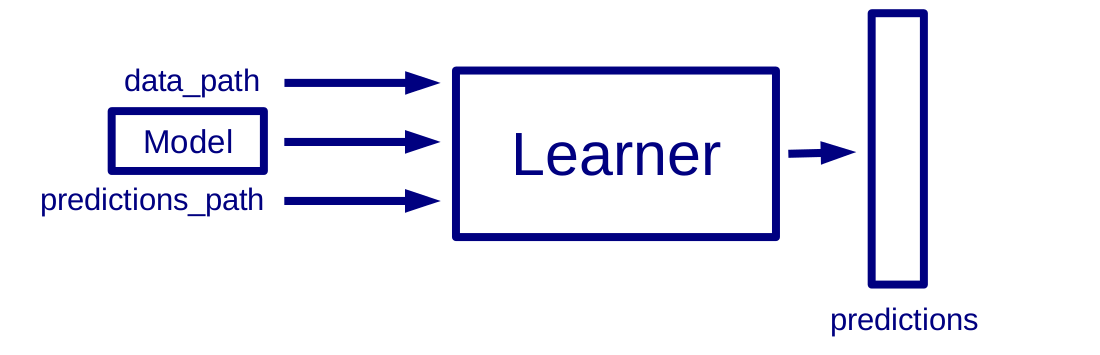
Inputs:
- path to a CSV file
- path to where the model is stored
- path to where the predictions will be stored
Outputs:
- predictions in CSV file (one label per line)
Example: the GPFunction learner produces several models. Let is pick the model called mostAccurate.txt and generate predictions for the same split higgs_noheader_02.csv.
$ java -jar gpfunction.jar -predict higgs_noheader_02.csv -model mostAccurate.txt -o predictions.csv
The executable gpfunction.jar will generate a csv file named predictions.csv containing one label per line.
Phase 2: Deployment during June
Once we have the final number of participants, each participant will be assigned a budget in Amazon EC2. Then, each participant will be asked to choose a combination of:
- EC2 flavor, i.e the virtual machine specs (check EC2 Instance Type Details here)
- running time per instance
- number of instances
- data-parallel strategy (% of data and % of variables/features sampled per instance)
In the last step prior to deployment, participants will have the option to expose a range of possible choices for their learner-specific parameters. This way, it will possible to assign different parameters to the different instances running on the cloud. More details to come on this aspect.
Finally, we will deploy your learners in EC2. We will analyze the predictions of your learner and communicate performance metrics.
The datasets:
In this edition of the workshop, we will target binary classification problems. For each dataset, we will release samples of different sizes. These samples will allow to estimate the running time of the classifier learning algorithms given the size of the data.
Higgs dataset:
The Higgs dataset is a public dataset. It is composed of 11000000 exemplars and 28 real-valued features. Expect a CSV file with any number of features where the last column is the label (0 or 1).
Blood Pressure Prediction dataset:
The BP dataset is a dataset composed of 4569200 exemplars. The goal is to predict acute hypotension episodes from physiological signals of thousands of patients. These signals are processed to identify beats and extract 100 real-valued features from those beats. Expect a CSV file with any number of features where the last column is the label (0 or 1).
Contact Map dataset:
The Contact Map dataset comes from the Protein Structure Prediction field, and it was originally generated to train a predictor for the residue-residue contact prediction track of the CASP9 competiton. The dataset has 32 million instances, 631 attributes, 2 classes, 98% of negative examples. For this activity, we will only consider the numerical features of this dataset (539 real-valued features). Expect a CSV file with any number of features where the last column is the label (0 or 1).
Support or Contact
Having trouble with the adaptation of your learner? Feel free to contact us by email at iarnaldo@mit.edu and we’ll try to help you sort it out.
Authors and Contributors
This collaborative activity is organized by the Any-Scale Learning For All (ALFA) group at MIT.
